Example Using the wine Dataset#
[1]:
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.metrics import f1_score
import featuristic as ft
import numpy as np
np.random.seed(8888)
print(ft.__version__)
1.0.1
Load the Data#
[2]:
X, y = ft.fetch_wine_dataset()
X.head()
[2]:
| alcohol | malicacid | ash | alcalinity_of_ash | magnesium | total_phenols | flavanoids | nonflavanoid_phenols | proanthocyanins | color_intensity | hue | 0d280_0d315_of_diluted_wines | proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 14.23 | 1.71 | 2.43 | 15.6 | 127 | 2.80 | 3.06 | 0.28 | 2.29 | 5.64 | 1.04 | 3.92 | 1065 |
| 1 | 13.20 | 1.78 | 2.14 | 11.2 | 100 | 2.65 | 2.76 | 0.26 | 1.28 | 4.38 | 1.05 | 3.40 | 1050 |
| 2 | 13.16 | 2.36 | 2.67 | 18.6 | 101 | 2.80 | 3.24 | 0.30 | 2.81 | 5.68 | 1.03 | 3.17 | 1185 |
| 3 | 14.37 | 1.95 | 2.50 | 16.8 | 113 | 3.85 | 3.49 | 0.24 | 2.18 | 7.80 | 0.86 | 3.45 | 1480 |
| 4 | 13.24 | 2.59 | 2.87 | 21.0 | 118 | 2.80 | 2.69 | 0.39 | 1.82 | 4.32 | 1.04 | 2.93 | 735 |
[3]:
y.head()
[3]:
0 1
1 1
2 1
3 1
4 1
Name: class, dtype: int64
Genetic Feature Synthesis#
[4]:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33)
synth = ft.GeneticFeatureSynthesis(
num_features=5,
population_size=200,
max_generations=100,
early_termination_iters=25,
parsimony_coefficient=0.05,
n_jobs=1,
)
synth.fit(X_train, y_train)
None
Creating new features...: 29%|████▋ | 29/100 [00:04<00:13, 5.27it/s]
Pruning feature space...: 100%|██████████████████| 5/5 [00:00<00:00, 551.52it/s]
Creating new features...: 29%|████▋ | 29/100 [00:05<00:12, 5.74it/s]
View the Synthesised Features and Their Formulas#
[5]:
generated_features = synth.transform(X_train)
generated_features.head()
[5]:
| alcohol | malicacid | ash | alcalinity_of_ash | magnesium | total_phenols | flavanoids | nonflavanoid_phenols | proanthocyanins | color_intensity | hue | 0d280_0d315_of_diluted_wines | proline | feature_24 | feature_3 | feature_0 | feature_1 | feature_10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 12.17 | 1.45 | 2.53 | 19.0 | 104 | 1.89 | 1.75 | 0.45 | 1.03 | 2.95 | 1.45 | 2.23 | 355 | -0.243548 | -19.029967 | 0.211763 | 0.249479 | -18.931961 |
| 1 | 13.87 | 1.90 | 2.80 | 19.4 | 107 | 2.95 | 2.97 | 0.37 | 1.76 | 4.50 | 1.25 | 3.40 | 915 | 0.008763 | -105.615575 | 0.016827 | 0.016909 | -89.365658 |
| 2 | 12.51 | 1.73 | 1.98 | 20.5 | 85 | 2.20 | 1.92 | 0.32 | 1.48 | 2.94 | 1.04 | 3.57 | 672 | -0.134808 | -29.173268 | 0.117513 | 0.136657 | -28.609145 |
| 3 | 13.88 | 1.89 | 2.59 | 15.0 | 101 | 3.25 | 3.56 | 0.17 | 1.70 | 5.43 | 0.88 | 3.56 | 1095 | 0.056938 | -92.415946 | -0.031179 | -0.032054 | -80.099472 |
| 4 | 13.40 | 3.91 | 2.48 | 23.0 | 102 | 1.80 | 0.75 | 0.43 | 1.41 | 7.30 | 0.70 | 1.56 | 750 | -0.564359 | 74.472843 | 0.512865 | 0.550195 | 67.989552 |
[6]:
info = synth.get_feature_info()
info
[6]:
| name | formula | fitness | |
|---|---|---|---|
| 0 | feature_24 | sin((sin((-(flavanoids) / cos((square(ash) / a... | -0.808449 |
| 1 | feature_3 | ((cos(flavanoids) + magnesium) - sin(abs(cos(p... | -0.821675 |
| 2 | feature_0 | sin((sin(sin(abs(-(flavanoids)))) / (flavanoid... | -0.822068 |
| 3 | feature_1 | sin((sin(flavanoids) / (flavanoids + 0d280_0d3... | -0.821911 |
| 4 | feature_10 | ((sin(cos(flavanoids)) + magnesium) - sin(abs(... | -0.819321 |
Feature Selection#
[7]:
def objective_function(X, y):
model = KNeighborsClassifier()
scores = cross_val_score(model, X, y, cv=3, scoring="f1_weighted")
return scores.mean() * -1
[8]:
selector = ft.GeneticFeatureSelector(
objective_function,
population_size=200,
max_generations=100,
early_termination_iters=25,
n_jobs=-1,
)
selector.fit(generated_features, y_train)
selected_features = selector.transform(generated_features)
Optimising feature selection...: 24%|██▏ | 24/100 [00:09<00:29, 2.56it/s]
View the Selected Features#
[9]:
selected_features.head()
[9]:
| alcohol | malicacid | ash | total_phenols | flavanoids | proanthocyanins | 0d280_0d315_of_diluted_wines | feature_24 | feature_1 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 12.17 | 1.45 | 2.53 | 1.89 | 1.75 | 1.03 | 2.23 | -0.243548 | 0.249479 |
| 1 | 13.87 | 1.90 | 2.80 | 2.95 | 2.97 | 1.76 | 3.40 | 0.008763 | 0.016909 |
| 2 | 12.51 | 1.73 | 1.98 | 2.20 | 1.92 | 1.48 | 3.57 | -0.134808 | 0.136657 |
| 3 | 13.88 | 1.89 | 2.59 | 3.25 | 3.56 | 1.70 | 3.56 | 0.056938 | -0.032054 |
| 4 | 13.40 | 3.91 | 2.48 | 1.80 | 0.75 | 1.41 | 1.56 | -0.564359 | 0.550195 |
Compare New Features to Original Features#
[10]:
model = KNeighborsClassifier()
model.fit(X_train, y_train)
preds = model.predict(X_test)
original_f1 = f1_score(y_test, preds, average="weighted")
original_f1
[10]:
0.7013084602631234
[11]:
model = KNeighborsClassifier()
model.fit(selected_features, y_train)
test_features = selector.transform(synth.transform(X_test))
preds = model.predict(test_features)
featuristic_f1 = f1_score(y_test, preds, average="weighted")
featuristic_f1
[11]:
0.965358758528083
[12]:
print(f"Original F1: {original_f1}")
print(f"Featuristic F1: {featuristic_f1}")
print(f"Improvement: {round(((featuristic_f1 / original_f1) - 1) * 100, 1)}%")
Original F1: 0.7013084602631234
Featuristic F1: 0.965358758528083
Improvement: 37.7%
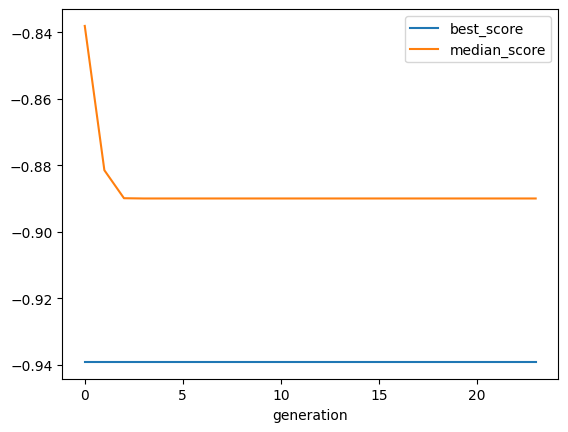
View the History of the Genetic Algorithms#
[13]:
synth.plot_history()
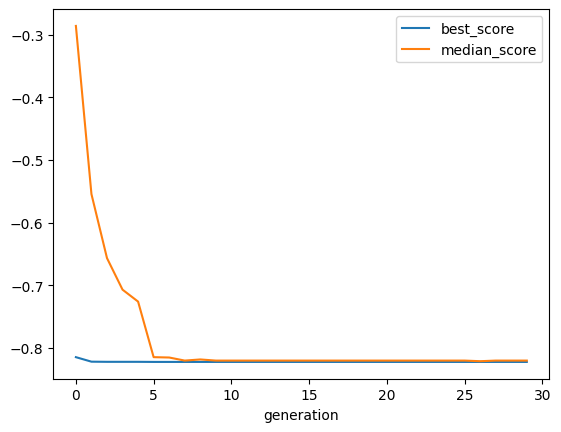
[14]:
selector.plot_history()