Using Featuristic With scikit-learn Pipelines#
Featuristic is compatible with scikit-learn’s powerful Pipeline class. This functionality allows you to organize and apply a sequence of data processing steps effortlessly within scikit-learn. With the Pipeline, you can chain together various transformers provided by Featuristic or other scikit-learn-compatible libraries.
These transformers can include feature generation, feature selection, data scaling, and any other preprocessing steps required to prepare your data for modeling.
By leveraging the Pipeline class in conjunction with Featuristic, you can streamline your data preprocessing workflow, ensuring consistency and reproducibility. This allows you to construct complex data processing pipelines with ease, and combine Featuristic with model development.
Let’s take a look at a simple example using the cars dataset.
[1]:
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import cross_val_score, train_test_split
from sklearn.pipeline import Pipeline
import featuristic as ft
import numpy as np
np.random.seed(8888)
print(ft.__version__)
1.0.1
Load the Data#
[2]:
X, y = ft.fetch_cars_dataset()
X.head()
[2]:
| displacement | cylinders | horsepower | weight | acceleration | model_year | origin | |
|---|---|---|---|---|---|---|---|
| 0 | 307.0 | 8 | 130.0 | 3504 | 12.0 | 70 | 1 |
| 1 | 350.0 | 8 | 165.0 | 3693 | 11.5 | 70 | 1 |
| 2 | 318.0 | 8 | 150.0 | 3436 | 11.0 | 70 | 1 |
| 3 | 304.0 | 8 | 150.0 | 3433 | 12.0 | 70 | 1 |
| 4 | 302.0 | 8 | 140.0 | 3449 | 10.5 | 70 | 1 |
[3]:
y.head()
[3]:
0 18.0
1 15.0
2 18.0
3 16.0
4 17.0
Name: mpg, dtype: float64
Split the Data in Train and Test#
[4]:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3)
Objective Function#
We define a custom objective function to pass into the Genetic Feature Selection algorithm. The output of this will be minimized to find the optimal subset of features.
[5]:
def objective(X, y):
model = LinearRegression()
scores = cross_val_score(model, X, y, cv=3, scoring="neg_mean_absolute_error")
return -scores.mean()
Fit a scikit-learn Pipeline Containing Featuristic#
[6]:
pipe = Pipeline(
steps=[
(
"genetic_feature_synthesis",
ft.GeneticFeatureSynthesis(
num_features=5,
population_size=200,
max_generations=100,
early_termination_iters=25,
parsimony_coefficient=0.035,
n_jobs=1,
),
),
(
"genetic_feature_selector",
ft.GeneticFeatureSelector(
objective,
population_size=200,
max_generations=50,
early_termination_iters=25,
n_jobs=-1,
),
),
(
"model",
LinearRegression()
)
]
)
model = pipe.fit(X_train, y_train)
Creating new features...: 74%|███████████▊ | 74/100 [00:15<00:05, 4.75it/s]
Pruning feature space...: 100%|██████████████████| 5/5 [00:00<00:00, 498.46it/s]
Creating new features...: 74%|███████████▊ | 74/100 [00:15<00:05, 4.74it/s]
Optimising feature selection...: 52%|█████▏ | 26/50 [00:05<00:05, 4.39it/s]
[7]:
preds = model.predict(X_test)
preds[:10]
[7]:
array([13.23930437, 35.40219719, 27.26035025, 26.25423477, 28.24773002,
19.10148319, 36.15024042, 23.33185658, 31.19121816, 22.29169501])
Accessing Featuristic Inside the Pipeline#
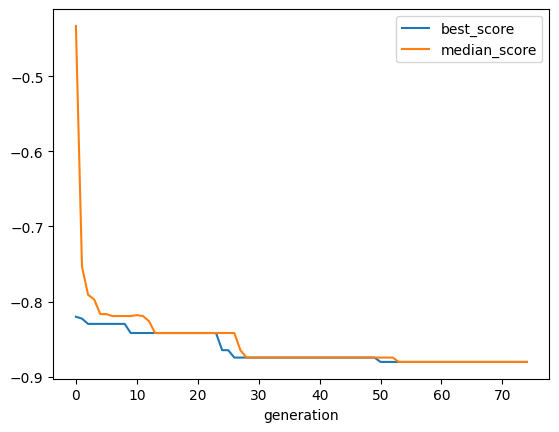
We can still access the individual Featuristic steps via the pipeline’s named_steps functionality. For example, to look at the formulas used for the feature engineering or to plot the genetic algorithm’s history.
[8]:
gfs = pipe.named_steps["genetic_feature_synthesis"]
gfs.get_feature_info().head()
[8]:
| name | formula | fitness | |
|---|---|---|---|
| 0 | feature_1 | abs(abs(abs((abs(abs(abs(abs(abs(abs(abs(abs(a... | -0.874683 |
| 1 | feature_8 | abs(abs(((cos(((abs(horsepower) / weight) - (s... | -0.847227 |
| 2 | feature_4 | abs(abs((abs(abs(abs(abs(abs(abs(abs(abs(abs(a... | -0.860441 |
| 3 | feature_5 | abs(abs(abs(abs(abs((((cube(horsepower) / (cub... | -0.852704 |
| 4 | feature_0 | abs(abs((model_year / abs(weight)))) | -0.880479 |
[9]:
gfs.plot_history()